
RxRx3-core
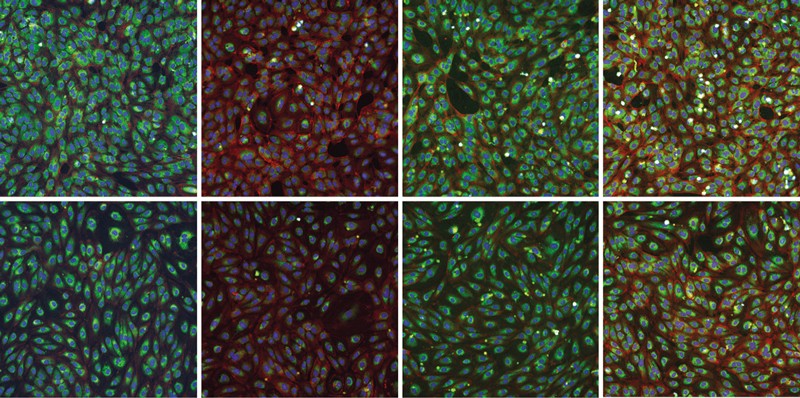
To accompany OpenPhenom-S/16, Recursion is releasing the RxRx3-core dataset, a challenge dataset in phenomics optimized for the research community. RxRx3-core includes labeled images of 735 genetic knockouts and 1,674 small-molecule perturbations drawn from the RxRx3 dataset and image embeddings computed with OpenPhenom-S/16. The dataset contains 6-channel Cell Painting images and associated embeddings from 222,601 wells but is less than 18Gb, making it incredibly accessible to the research community.
Mapping the mechanisms by which drugs exert their actions is an important challenge in advancing the use of high-dimensional biological data like phenomics. We are excited to release the first dataset of this scale probing concentration-response along with a benchmark and model to enable the research community to rapidly advance this space.
Paper published at LMRL Workshop at ICLR 2025: RxRx3-core: Benchmarking drug-target interactions in High-Content Microscopy.
Building Maps of Biology and Chemistry
At Recursion, we build maps of biology and chemistry to explore uncharted areas of disease biology, unravel its complexity, and industrialize drug discovery. Just as a map helps to navigate the physical world, our maps are designed to help us understand as much as we can about the connectedness of human biology so we can navigate the path to new medicines more efficiently.
Our maps are built using image-based high-dimensional data generated in-house. We conduct up to 2.2 million experiments every week in our highly automated labs, where we use deep learning models to embed high dimensional representations of billions of images of human cells that have been manipulated by CRISPR/Cas9-mediated gene knockouts, compounds, or other reagents. This allows us to create representations that can be compared and contrasted to predict trillions of relationships across biology and chemistry — even without physically testing all of the possible combinations. Recursion's Maps and associated applications help navigate complex biology and chemistry by revealing relationships across genes and chemical compounds.
Map Benchmarking
Test your maps with the rxrx-compound-gene-activity-benchmark benchmark!