Background
Target details
Epidermal Growth Factor Receptor (EGFR) is a transmembrane protein that plays a critical role in cell growth, differentiation, and survival. It is frequently overexpressed or mutated in various cancers, including non-small cell lung cancer, colorectal cancer, and head and neck cancer. This makes EGFR a crucial target for cancer therapies such as Cetuximab, an antibody with more than 1B USD in annual revenue.
- Target Protein: EGFR
- Organism: HUMAN
- Uniprot Accession ID: P00533
- Protein sequence: LEEKKVCQGTSNKLTQLGTFEDHFLSLQRMFNNCEVVLGNLEITYVQRNYDLSFLKTIQEVAGYVLIALNTVERIPLENLQIIRGNMYYENSYALAVLSNYDANKTGLKELPMRNLQEILHGAVRFSNNPALCNVESIQWRDIVSSDFLSNMSMDFQNHLGSCQKCDPSCPNGSCWGAGEENCQKLTKIICAQQCSGRCRGKSPSDCCHNQCAAGCTGPRESDCLVCRKFRDEATCKDTCPPLMLYNPTTYQMDVNPEGKYSFGATCVKKCPRNYVVTDHGSCVRACGADSYEMEEDGVRKCKKCEGPCRKVCNGIGIGEFKDSLSINATNIKHFKNCTSISGDLHILPVAFRGDSFTHTPPLDPQELDILKTVKEITGFLLIQAWPENRTDLHAFENLEIIRGRTKQHGQFSLAVVSLNITSLGLRSLKEISDGDVIISGNKNLCYANTINWKKLFGTSGQKTKIISNRGENSCKATGQVCHALCSPEGCWGPEPRDCVSCRNVSRGRECVDKCNLLEGEPREFVENSECIQCHPECLPQAMNITCTGRGPDNCIQCAHYIDGPHCVKTCPAGVMGENNTLVWKYADAGHVCHLCHPNCTYGCTGPGLEGCPTNGPKIPS
- Structure PDB: 6ARU
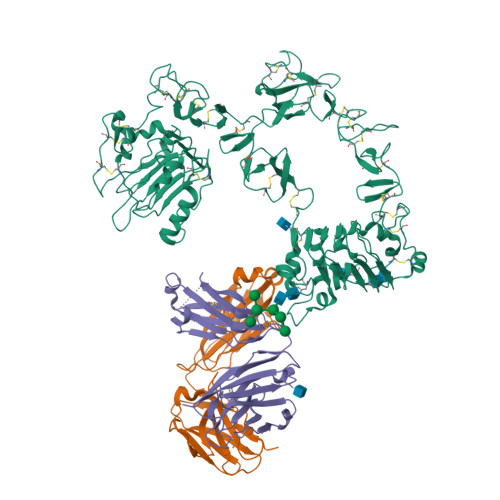
Binding protein designs
This dataset contains 202 designed EGFR-binding protein sequences, along with experimental binding affinity results tested by the AdaptyvBio team, plus 11 additional sequences ordered by Anthony Gitter and tested by the AdaptyvBio team.
Benchmark description
This retrospective benchmark evaluates protein design methods by challenge participants to design a binding protein for the extracellular domain of EGFR, a cancer-associated drug target. A set of 202 previously designed protein sequences, along with their experimental binding affinities (binary labels), is available for testing.
Balanced accuracy is used to evaluate the performance of design methods in differentiating between binders and non-binders.
Additional notes
Cetuximab_scFv and P01133-971-1023 were used as the positive control of the binding assay.ahmedsameh-Q3 and ahmedsameh-yy2 were disqualified from the competition due to these similarity levels to the known EGFR-binder sequence. They are kpet in this benchmark solely for evaluation purposes.- The two weak binders
alecl-Sequence1 and alan.blakely-design:5 n:6|mpnn:1.247|plddt:0.825|ptm:0.709|pae:10.151|rmsd:3.535 are classified as non-binders in this benchmark.
Reference: